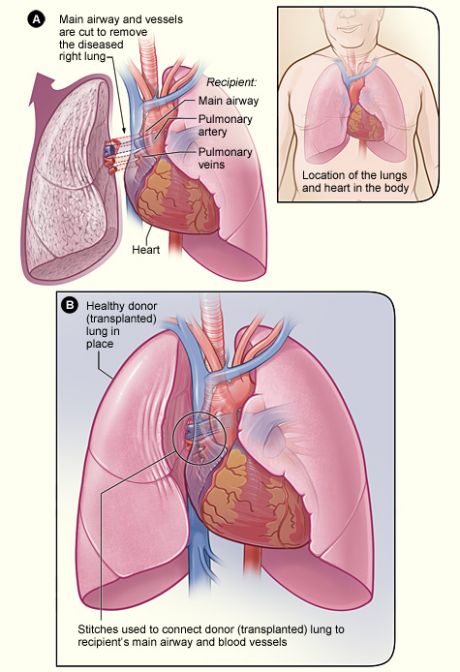
LT is often the only treatment option for patients with chronic respiratory failure. However, LT procedures are associated with chronic lung allograft dysfunction (CLAD), a condition that presents with obstructive and restrictive ventilation in nearly 50 % of LT patients. This requires administration of immunosuppressant drugs or even re-transplantation.
An early prediction of CLAD would facilitate the implementation of novel interventions to improve disease outcome. In this context, the EU-funded SYSCLAD (Systems prediction of chronic lung allograft dysfunction) project set out to identify a clinical and molecular signature profile predictive of CLAD and a computational model of disease mechanisms for interpreting and validating the molecular data. This would speed up diagnosis and allow for early intervention before significant organ damage ensues.
Over 400 patients with LT were recruited and their samples analysed by transcriptomics, proteomics and sequencing at selected time points. The data was integrated into a mathematical model alongside clinical information, leading to the identification of signatures of each CLAD sub-phenotypes. These phenotypes were based on differential gene and protein expression as well as specific patterns of pulmonary microbiota composition and alveolar inflammation.
SYSCLAD combined systems biology and clinical events to successfully model the full spectrum of CLAD mechanisms. Their computational model is capable of interpreting molecular and clinical information to accurately predict LT recipients at risk of developing CLAD three years post-transplantation. Given the poor median survival figures from the time of CLAD diagnosis, this medical tool should expedite intervention and improve LT outcome before lung function is irreversibly lost.