Elucidating the structure of intra-membrane proteases
Elucidating the structure of intra-membrane proteases
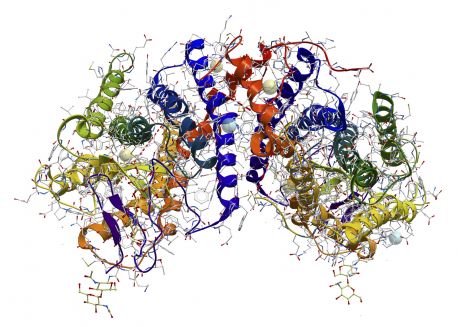
The three-dimensional structure of proteins is central to understanding their function. European researchers focused on the crystallisation of membrane-bound enzymes.
Membrane proteins represent one third of the proteins of living
organisms. Although half of the drug targets are membrane proteins, only
1 % of these have been structurally characterised. Presenilin is an
intramembrane protease involved in several physiological processes and
in the early onset of Alzheimer’s disease.
Prior efforts to determine the structure of the human protein presenilin had failed. For this reason, scientists resorted to studying their prokaryotic preselinin homologues (PSH) under the aegis of the EU-funded INTRAMEMPROT (Mechanistic and structural insight into di-aspartyl intramembrane proteases) project. They worked to gain structural and functional insight into a family of membrane proteins from archaea as they are related in structure to presenilin.
They cloned 33 different PSHs as well as enzyme-stabilising mutants for expression in E.coli. Scientists successfully produced and purified ten of these PSH clones. However, despite extensive optimisation efforts to obtain good crystals of PSH, insufficient X-ray diffraction prohibited the structural investigation of PSH.
As a result, the consortium switched its efforts towards the structural determination of CDP-alcohol phosphotransferases. These are membrane proteins responsible for the biosynthesis of fundamental membrane lipids. One such lipid, cardiolipin, is essential for respiratory processes in bacteria and eukaryotic mitochondria. The structure of a CDP-alcohol phosphotransferase from archaea was determined, providing for the first time insight into the reaction mechanism of this family of membrane-embedded enzymes.
Collectively, the activities of the INTRAMEMPROT project successfully overcame general limitations associated with the structure determination of membrane-bound enzymes. The generated information has led to important conclusions regarding their substrate-binding domains and conformational changes. Finally, the adapted methodology could extend to the crystallisation of other proteins.
published: 2016-07-08